Project B7
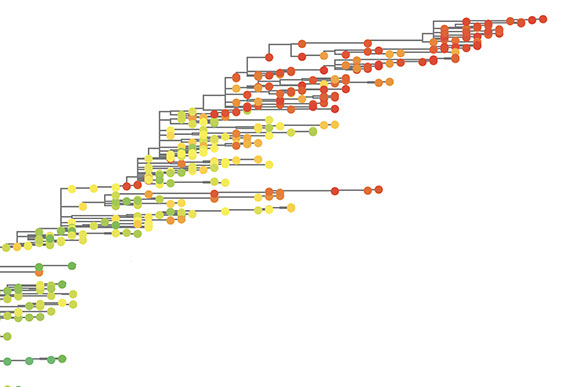
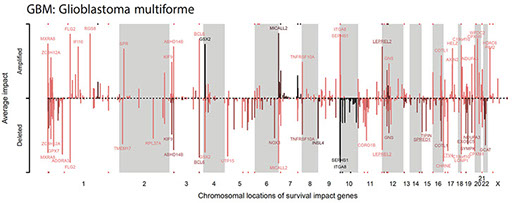
Predicting the constrained evolution of tumors
Andreas Beyer, U Cologne | web | email
This project takes a systems-biology modelling approach to tumor evolution, based on tissue-specific genomic and epigenetic factors influencing mutation rates. Combined with selection estimates from molecular network and pathway data, this project will predict the tissue-specific likelihood of mutations driving tumor evolution.
Predictability in Evolution
Collaborative Research Center 1310
Publications
Convergent network effects along the axis of gene expression during prostate cancer progression
Charmpi, K., Guo, T., Zhong, Q. et al., Beyer A., Genome Biol 21, 302, 14. December 2020, https://doi.org/10.1186/s13059-020-02188-9
Kamrad S., Grossbach J., Rodríguez‐López M., Mülleder M., Townsend S.J, Cappelletti V., Stojanovski G., Correia‐Melo C., Picotti P., Beyer A., Ralser M., Bähler J., Mol Syst Biol (2020)16:e9270, 1. April 2020, https://doi.org/10.15252/msb.20199270
Multi-region proteome analysis quantifies spatial heterogeneity of prostate tissue biomarkers
Guo T., Li L., Zhong Q., Rupp N.J., Charmpi K., Wong C.E., Wagner U., Rueschoff J.H., Jochum W., Fankhauser C.D., Saba K., Poyet C., Wild P.J., Aebersold R., Beyer A., Life Science Alliance, 29. May 2018, https://doi.org/10.26508/lsa.201800042
regNet: an R package for network-based propagation of gene expression alterations
Seifert M., Beyer A., Bioinformatics Volume 34 Issue 2:308–311, 15. January 2018, https://doi.org/10.1093/bioinformatics/btx544
Christodoulou E.G., Yang H., Lademann F., Pilarsky C., Beyer A., Schroeder M., Oncotarget 8:34283-34297, 10. March 2017, https://doi.org/10.18632/oncotarget.16079